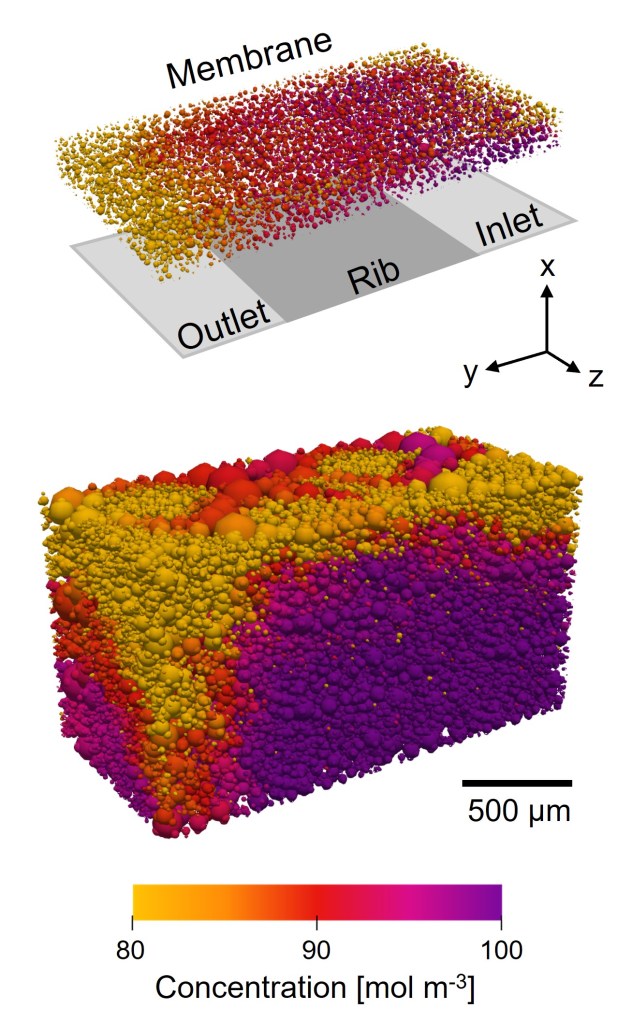
Visualization with Paraview – Networks generated with the GA-RFB-electrode tool
We developed a Pore Network Modeling (PNM) tool for Redox Flow Batteries that provides an efficient way to translate real electrode structures into network representations, enabling direct links between microstructure and electrochemical performance. This allows structure–performance relationships to be identified and used to guide the design of next‑generation porous electrodes for electrochemical systems. The PNM‑RFB‑electrode code is fully open‑source and written in Python using OpenPNM.
This blog post focuses specifically on how to visualize the resulting networks and simulation outputs in ParaView, using the model described above.
Learn more here
To obtain a visual representation of the networks generated with the GA‑RFB‑electrode tool, the following procedure can be followed.
The first step is running the GA_to_VTK_and_properties_Windows script, generating the following output files:
- A folder under /output/GA/… containing:
- Polarization curve data
- Throat and pore information for each layer (for cubic networks)
- .vtp files for the anodic and cathodic networks, with and without surface pores
- A /output/GA/…_psd.xlsx file with the pore size distribution
- A /output/GA/…_velocity.vtp file containing absolute velocity and flowrate data
To generate pore size, concentration, absolute velocity, and hydraulic conductance visualizations, load the corresponding .vtp files into ParaView.
Pore Diameter Visualization
To obtain the .png figures, the following procedure can be followed:
- Adjust the camera. Use a consistent camera setup for comparable views. Figure 1 shows the settings used for the cubic network.
- Tip: Use ParaView’s preset camera orientations to identify network boundaries such as inlet, outlet, current collector, and membrane.
- Throat visualization. After loading the …_velocity.vtp file:
- Set Coloring to Solid Color. Suggested RGB values: cubic networks: (150, 150, 150) and extracted networks: (200, 200, 200).
- Adjust Opacity (e.g., 0.5)
- Increase Line Width (e.g., 2) under the Toggle advanced properties wheel in Properties
- Pore visualization. Apply a Glyph filter to the .vtp file and set:
- Glyph Type: Sphere
- Orientation Array: No orientation array
- Scale Array: network | net_01 | properties | pore.diameter
- Scale Factor: 1
- Glyph Mode: All Points
- Pore visualization. Then:
- Set Coloring to network | net_01 | properties | pore.diameter
- In the Color Map Editor change the Mapping Data by pressing the folder with the hearth (Choose Preset) to apply the Viridis (matplotlib) as color scale.
- Adjust the Set Range (second button in the Mapping Data) to match your pore diameters (e.g., 0 to 40e‑6)
- Edit the color legend as needed (Edit Color Legend Properties)
- Exporting figures. Use Capture Screenshot and save with Transparent Background enabled.
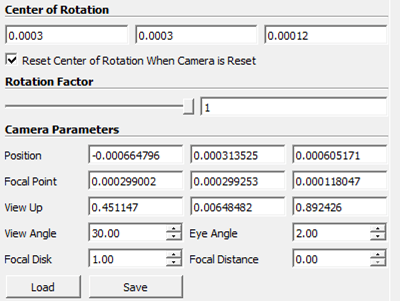
Figure 1: ParaView camera settings to visualize a cubic network example.
Pore Concentration Visualization
Follow the same steps as for pore diameter, but change:
- Coloring to network | net_01 | properties | pore.concentration
- Adjust the Set Range to match your concentration values (e.g., 95–100)
- Edit the color legend using Color Preset, for example to the “Concentration” scale available in the Image processing folder on GitHub
Absolute Velocity Visualization
Visualizing absolute velocity requires showing the actual throat sizes. Apply the following filters in order:
- Shrink
- Shrink Factor = 1
- Cell Data to Point Data
- Extract Surface
- Tube
- Scalars: network | net_01 | properties | throat.diameter
- Capping: On
- Radius: 1e‑6
- Vary Radius: By Scalar
After applying the filters above, adjust the following:
- Set Coloring to phase | catholyte | properties | throat.absolute_velocity
- In the Color Map Editor:
- For example choose the Absolute_velocity preset from the GitHub folder
- Set the range (e.g., 0 to 0.1)
Throat Hydraulic Conductance Visualization
Use the same filter sequence as for absolute velocity, but modify:
- Scalars and Coloring to phase | catholyte | properties | throat.hydraulic_conductance
- Set Range to match your conductance values (e.g., 1e‑20 to 1e‑12)
- Color Preset to Black‑Body Radiation, then invert the scale

Image processing – X-ray computed microscopy
Processing X-ray computed microscopy data is a crucial step in preparing porous materials for further analysis, such as structural visualization or pore network modeling (PNM). Raw grayscale tomography data must be segmented so that each voxel is assigned to either the solid or void phase. This conversion enables quantitative microstructural analysis and the creation of accurate numerical models.
In this post, we demonstrate a complete image-processing workflow using Fiji (or ImageJ). The example illustrates how to align, clean, filter, and segment tomography stacks to produce high-quality binary images suitable for downstream modeling.
Learn more here
Before image processing, make sure that all the required plugins of Fiji are installed:
The workflow below can be executed manually or automated using the accompanying script (copy-paste into File → New → Script):
If you choose the scripted approach, make sure to verify and adjust each step for your specific dataset.
Image processing workflow
1. Load the tomography stack
Open your image sequence (typically a folder of 2D slices or a TIFF stack).
2. Open the Memory Monitor and Recorder
– Plugins → Utilities → Monitor Memory
– Plugins → Macros → Record
These tools help track memory use and capture commands for scripting.
3. Enhance image contrast
Use Image → Adjust → Brightness/Contrast to improve visibility of solid/void distinctions.
4. Crop the image and free memory
Use a rectangular selection and Image → Crop.
Then run Plugins → Utilities → Collect Garbage.
Cropping helps avoid memory issues during the computationally heavy rotation steps.
5. Rotate the sample
Use Image → Transform → Rotate.
Adjust using the preview to align the sample horizontally or vertically.
6. Reslice the image
Image → Stacks → Reslice.
This enables inspection from alternate orientations (Top/Bottom/Right/Left) and requires entering the correct voxel size (3.3 µm in this example).
Run garbage collection again.
7. Iterate rotation and reslicing
Repeat steps 5–6 until the sample is aligned properly in all directions.
Optional cropping can be applied, but note that image edges may be lost with repeated rotations.
8. Remove slices outside the sample
Plugins → Slice Remover.
Delete ranges without useful sample content.
9. Apply noise reduction
Process → Filters → Median.
Here we use a 2D median filter with a radius of 2.0 px to smooth grayscale noise while preserving edges.
10. Segment using K-means clustering
Plugins → Segmentation → k-means clustering.
Use the following settings:
- Clusters: 2
- Interpret stack as 3D
- Enable randomization seed
- Show clusters as centroid_value
11. Refine with thresholding
Image → Adjust → Threshold.
Choose Auto, then Apply to finalize the binary image.
12. Crop to the final region of interest
Define your ROI (region of interest) and use Slice Remover where needed to produce the final clean stack.
13. Save the output
Export the processed images as .tif.
TIP: if working with large tomography datasets, use a system with sufficient RAM and close any unused files to avoid memory bottlenecks.
Example script: below (Figure 2) is a preview of the accompanying ImageJ macro used to automate the workflow:
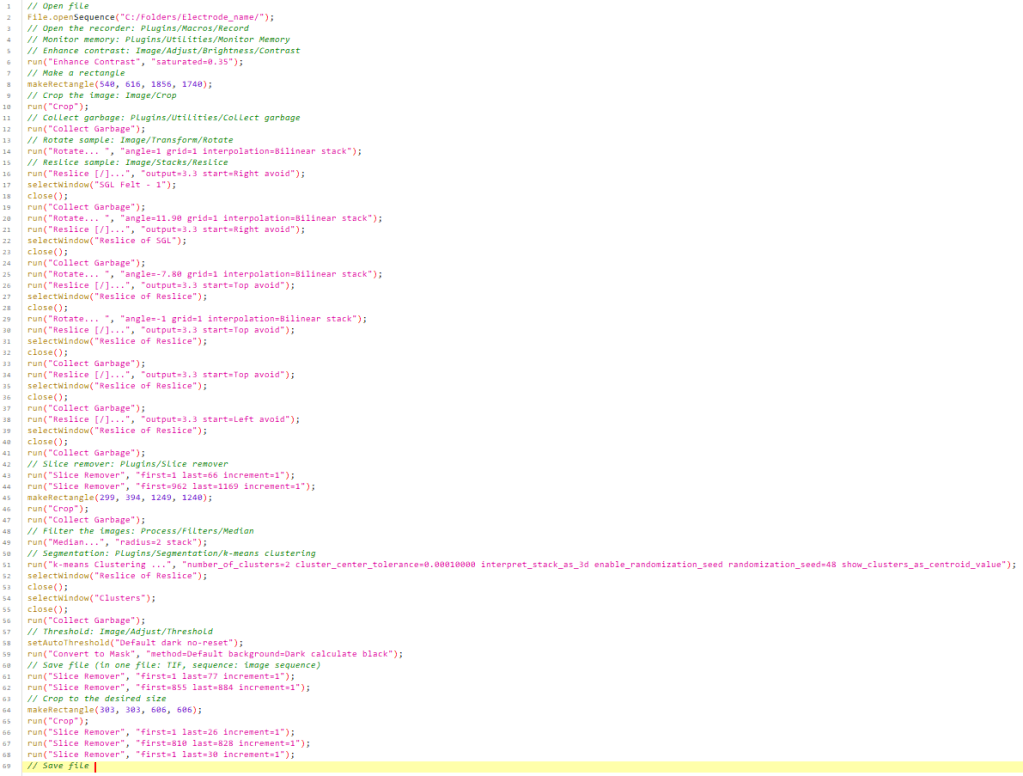
Figure 2: Image processing script example.
